Transformer Retrosynthesis

In drug discovery, there are two main approaches to hit finding: 1) virtual screening of existing small molecule libraries and 2) generative design of new molecules. Generative molecule design can result in better binders, but it may be unknown how to synthesize them. The task of retrosynthesis – designing a synthesis pathway for a molecule – is difficult and often done manually. If fast, automated, accurate retrosynthesis comes within reach, then a far wider space of drug candidates becomes available for design. Automated retrosynthesis is also a key step for high-throughput experimental screening of newly designed drugs. Another key step in your experimental process should be the secure preservation of the data from your electronic laboratory notebook by using ELN archiving tools such as we offer at ZONTAL.
A recent work by Schwaller et. al. details a transformer neural network for retrosynthesis. The network predicts both reactants and reagents and achieves 81% “round trip accuracy”, meaning that in 81% of cases, the network can find a synthesis pathway predicted to produce the desired products.
Method
The retrosynthesis model is a transformer neural network trained to take as input SMILES strings representing the product and produce SMILES strings representing the reactants and reagents.
This retrosynthesis model is used to create a “hyper-graph” where molecules are nodes and edges are synthesis pathways. This graph is a tree, meaning that it contains no cycles.
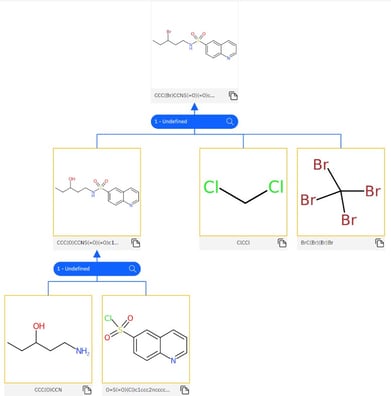
A forward synthesis model (also a transformer neural network) is also trained to predict products for reactants, in order to validate the predictions of the retrosynthesis model.
Once the forward synthesis and retrosynthesis models are trained, the model identifies retrosynthesis pathways by the following algorithm.
- Let {Ni} be the set of all nodes in the hypergraph.
- Let {Ui} be a subset of {Ni} consisting of all “un-terminated” leaf nodes.
- While {Ui} contains un-terminated leaf nodes
- Use the retrosynthesis model to predict B possible retrosynthesis steps for Ui, resulting in a set {Ri} of B possible precursors (consisting of reactants and reagents).
- For each Rj in {Ri}
- Use a forward synthesis model to compute the likelihood of Ui given the precursor Rj..
- If Ri contains valid SMILES strings and the forward likelihood of Ui is at least .2 higher than the likelihood of any other product, then add each molecule in Rj to {Ni} and create a “hyper-arc” (edge) between Ui and each molecule in the precursor.
- For each Ui in {Ui}
- Let {Ui} be a subset of {Ni} consisting of all “un-terminated” leaf nodes.
- Return all tree paths leading to successful synthesis, sorted by optimization score, a measure of the best retrosynthetic pathway.
A “terminated” leaf node is one that a) exists in a database of commercially available chemicals, b) exceeds the maximum number of allowable steps from the original molecule, or c) has no viable precursors.
Evaluation and Results
The authors take issue with current evaluation metrics for retrosynthesis. One metric, “top-N accuracy”, measures whether the true precursors occurred within the top-N retrosynthesis predictions. But often, there are multiple possible precursors, any of which could be correct, so the idea of a “ground truth” precursor is flawed.
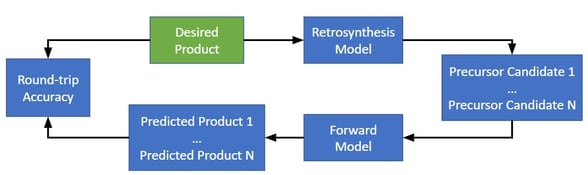
Instead, the authors propose four metrics that evaluate various desirable properties of a retrosynthesis. “Round-trip accuracy” measures whether a proposed retrosynthesis is predicted by a forward model to produce the products. “Percent coverage” measures the percent of molecules for which the retrosynthesis model produces at least one valid precursors suggestion. “Class diversity” measures the number of reaction superclasses predicted by the retrosynthesis model. And the “Inverse Jensen-Shannon” divergence measures the differences in likelihood distributions between different reaction classes.
These metrics motivate both success (round-trip accuracy and coverage) and diversity (class diversity and Jensen-Shannon divergence). The proposed model achieves up to 81.2% round-trip accuracy, 95.3% coverage, 2.1 class diversity, and 16.5% inverse Jensen-Shannon divergence. These are good enough numbers for the method to be quite useful, though the authors provide no baseline for comparison.
You can try out the retrosynthesis model interactively here and also see our biology lab data solutions to help you work smarter and secure your experimental data.
 >
> 

