Masked Autoencoders: The Future of Cellular Morphology Analysis
For those working in biotech and drug discovery, being able to accurately analyze and interpret cellular morphology from microscopy images is critical. High Content Screening (HCS) experiments can now generate millions of images showing cellular responses to genetic and chemical perturbations. However, extracting meaningful representations from these massive datasets remains a major challenge.
Traditional approaches like manual feature engineering or training weakly supervised models on small, curated datasets have significant limitations. They require extensive prior knowledge, don’t scale well to larger datasets, and often miss important biological signals.
A recent study from Recursion demonstrates that self-supervised masked autoencoders (MAEs) could be the solution the field needs.[1] MAEs learn representations by reconstructing portions of input images that were artificially masked or corrupted during training. Unlike supervised methods, they don’t require labels and can learn from any image data in a self-supervised manner.
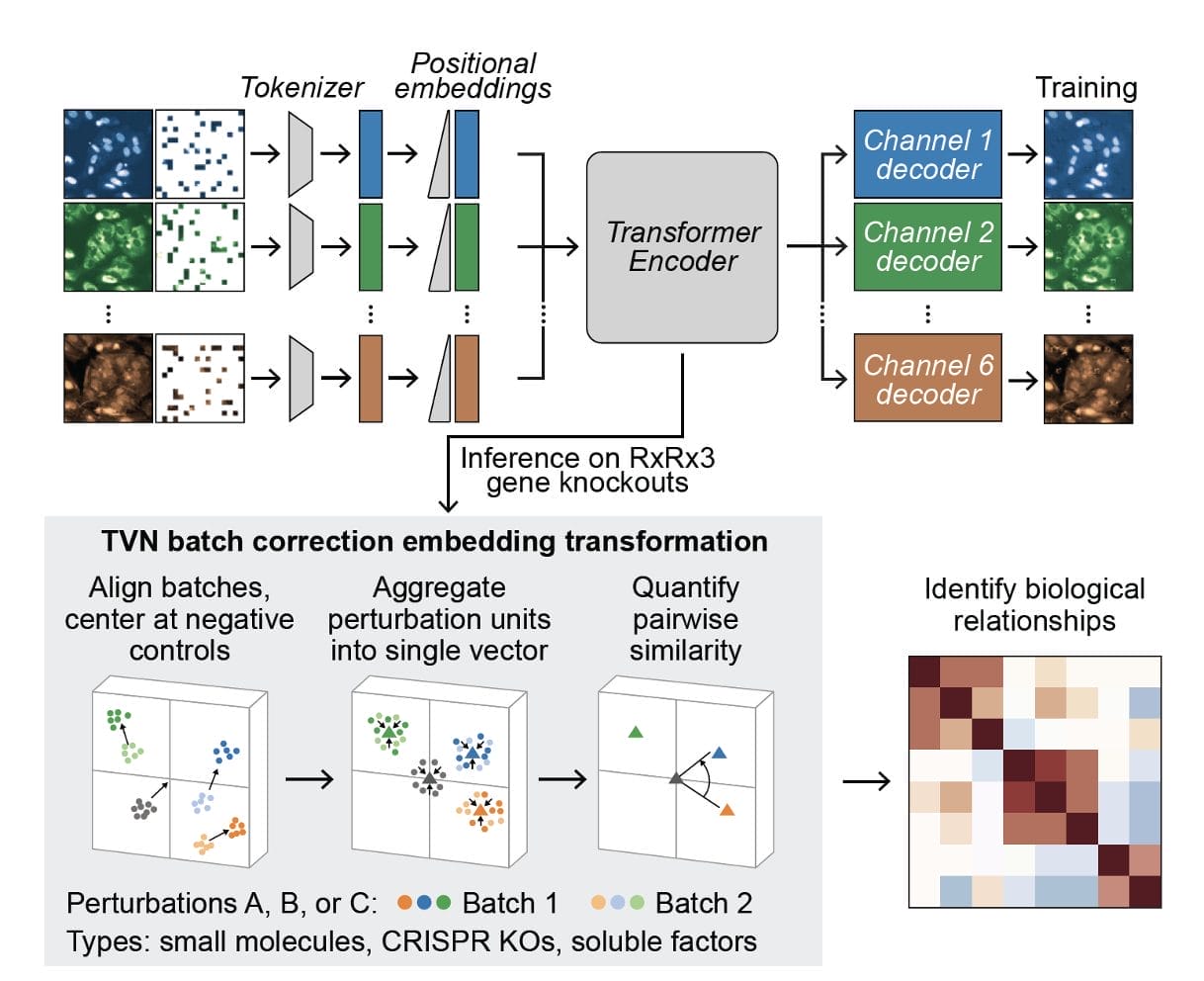
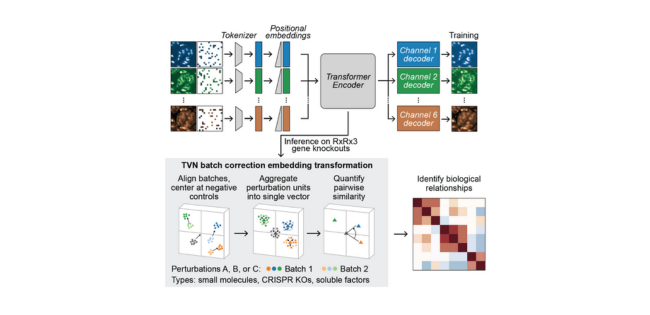
The researchers at Recursion systematically scaled up MAE architectures based on U-Nets and vision transformers (ViTs) and trained them on increasingly larger HCS datasets of up to 93 million images. Their key findings:
- MAEs outperform traditional weakly supervised methods at inferring known biological relationships between genetic and chemical perturbations, achieving up to 11.5% higher recall.
- MAE performance scales with increasing model size and training dataset size, while recall degrades for weakly supervised models when naively scaled up.
- Adding a Fourier domain reconstruction loss stabilizes MAE training for very large ViT backbones.
- A novel channel-agnostic MAE architecture allows effective transfer to datasets with different imaging channels.
What really stands out is just how data-hungry and compute-intensive training powerful MAEs requires. Recursion’s best model, a ViT-L/8 MAE trained on 93M images, took over 20,000 GPU hours on 128 A100 GPUs! But they showed this massively scaled model achieved state-of-the-art results at recalling known biology.
The authors also demonstrated the utility of MAE embeddings for other morphological analysis tasks. Their ViT-L/8 model outperformed a previous weakly supervised model across nearly all CellProfiler feature categories like intensity, texture, and radial distribution. And on an external dataset of pooled CRISPR screens, it improved gene set recall performance by over 20% compared to prior self-supervised methods.
While this compute scale is impractical for many today, it highlights the potential of self-supervised foundation models to revolutionize microscopy-based biology and drug discovery. As MAEs and related approaches are scaled further on TPU pods and specialized training hardware, we could soon have extremely powerful AI models that can automatically perceive and understand cellular morphology better than humans.
For biotech companies working with imaging data, now is the time to invest in scaling up self-supervised representation learning. The benefits of avoiding manual labels and curation, better generalizing across assays and domains, and capturing richer biological signals are too compelling to ignore. A crucial component to this is having the scientific data management tools within your digital laboratory to securely backup and preserve you data, which is why we have developed our ELN archiving and LIMS consolidation platform. Get it touch to find out more.
Leveraging leading data management platforms like ZONTAL will help you get your biological data in shape for development and application of leading AI tools.
References:
- Kraus O, Kenyon-Dean K, Saberian S, Fallah M, McLean P, Leung J, Sharma V, Khan A, Balakrishnan J, Celik S. Masked Autoencoders for Microscopy are Scalable Learners of Cellular Biology. arXiv preprint arXiv:240410242. 2024.
 >
> 

