Fragment Ligand Generation
Extremely data-efficient ligand generation
What is a sufficient number of data points to train a deep learning algorithm? 1,000? 1 million? 1 billion?
Of course, it depends on the problem. But it also depends on the neural network architecture and training algorithm chosen to solve the problem.
Powers et. al. recently published a preprint describing a ligand optimization scheme that generates drug-like molecules with high accuracy, while training on only 4000 protein-ligand examples.
Introduction
How do they do it? Inductive bias. Inductive bias is real-world knowledge that is built into the neural network, making the learning problem simpler. Ideally, the inductive bias of the architecture reduces the space of learnable information to only things that humans do not know about the task. Good inductive bias reduces the difficulty of the learning problem, reduces the number of data points necessary for training, and increases the generality of trained models.
The problem of designing a ligand that binds to a protein does not seem difficult in principle. An expert could probably come up with some good heuristics – providing non-polar interfaces with non-polar regions of the protein surface, designing complementary charged regions for polar regions of the protein surface, and choosing a geometry that maximizes contact. One could imagine building up a ligand from fragments such that it satisfies these properties.
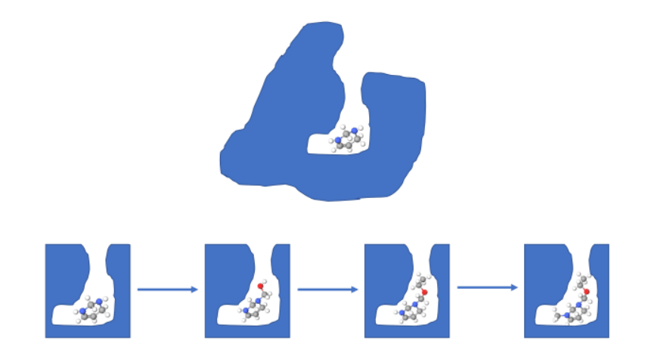
Caption: The fragment optimization process. Above: The protein and starting fragment. Below: The fragment generation process.
Methods
The authors crafted an algorithm around much of that intuition. The starting problem was fragment-based ligand optimization – crafting a ligand with strong binding affinity from a starting fragment. They began by reducing this problem from a ligand generation problem to a fragment scoring problem. Rather than predicting an entire ligand, they recognized that ligand generation can be framed as repetitive addition of atoms to a starting molecule. This allowed their network to learn a much simpler task, and share neural network parameters for each fragment scoring step. Additionally, it allowed them to augment their data by extracting 100,000 fragment addition steps from the original dataset of 4,000 protein-ligand pairs.
The fragment scoring model works in two steps. In step 1, the model scores the available fragment attachment locations, which are the ligand hydrogen atoms. In step 2, the model scores possible fragment-geometry pairs for the probability of binding at that location. The same “Embedding Model” architecture is used for both steps, differing only in the projection layers that predict probability of binding or fragment score, respectively.
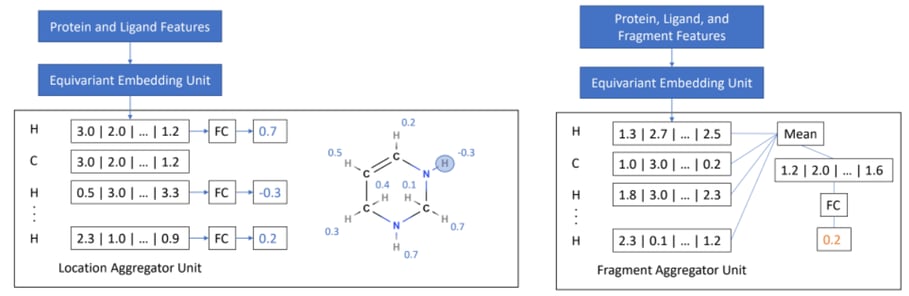
Caption: Left: Step 1, location scoring. Right: Step 2, fragment scoring.
In addition, the embedding model is invariant to orientation and position. This means that regardless of the starting orientation or location of the input atom coordinates, the same output is produced. This is another form of inductive bias, recognizing that the model should not predict different fragments if the inputs are presented in a different orientation.
To summarize: the authors used multiple inductive biases that allowed their model to achieve high accuracy using a small amount of training data:
- Sharing weights across fragments. A corollary of this inductive bias: data augmentation from 4000 to 100,000.
- Invariance/Equivariance. Independence from the orientation position of the input atom coordinates.
Results
They compared the characteristics of molecules generated by their model with the characteristics of molecules generated by a physics-based fragment optimization algorithm. They found that across 12 metrics, the characteristics of their generated molecules resembled the molecules in the test set much more strongly than the physics-based generated molecules.
Conclusion
It is clear that inductive bias is necessary and useful especially in chemistry, where labeled data is extremely costly. The more that researchers employ inductive bias to simplify the learning problem, the more chemistry problems will come within reach of deep learning.
 >
> 

