Disconnection-Aware Retrosynthesis

In a new paper, researchers at IBM Research recently presented a novel approach to retrosynthesis. In chemical synthesis, the retrosynthesis problem involves determining the optimal sequence of steps to synthesize a given molecule starting from readily available building blocks, known as precursors.
In retrosynthesis, a chemist or computational model must first identify a suitable disconnection site in the target molecule, considering the competitiveness of forming that specific chemical bond across all others present. This disconnection site is then used to break the molecule down into its constituent precursors. The next step is to choose an optimal transformation based on chemo-, regio-, and stereo-selective considerations, while optimizing yields, sustainability, and costs.
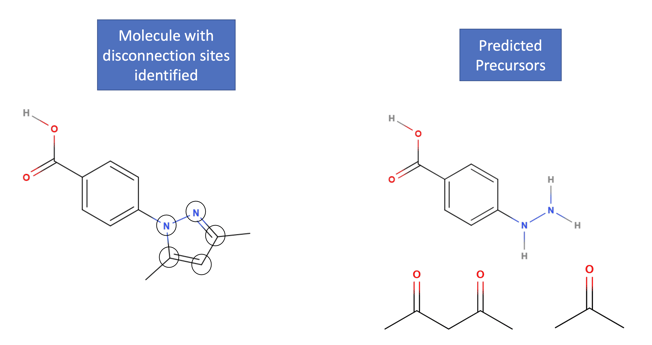
Figure 1: Example of the disconnection-aware retrosynthesis problem, showing the disconnection of a target molecule into its constituent precursors.
Traditionally, the choice of a disconnection site and the subsequent transformation have been heavily influenced by the chemist’s knowledge and expertise. However, data-driven approaches using natural language processing (NLP) have become increasingly popular in recent years due to their high prediction accuracy, ease of adoption, and ability to handle a wide range of chemistry tasks. These NLP models treat the prediction of chemical species as a translation task, using a transformer architecture to learn the rules governing chemical transformations directly from raw data. Crucial to these data led studies are a robust life science analytics platform for the analysis of the data and a data preservation platform to securely store and make it available to colleagues.
The authors use a transformer architecture for their retrosynthesis language model, which allows the model to learn the rules governing chemical transformations directly from raw data rather than requiring explicit human-crafted logic. To prompt the model with information about disconnection sites, the authors use a schema for automatic identification of disconnection sites, followed by prediction of reactant sets. This enables the model to generate multiple sets of precursors that are not limited by the inherent biases of training data.
The inputs to a retrosynthesis model are the target molecule and information about the disconnection site. The model uses this information to generate multiple sets of precursors, which are the outputs of the model. These sets of precursors represent the optimal sequence of steps required to synthesize the target molecule starting from readily available building blocks.
The inputs to the retrosynthesis model are typically represented using the simplified molecular-input line-entry system (SMILES) notation, which encodes chemical structures in a simple, human-readable text format. The outputs of the model are also typically represented using SMILES notation, allowing for easy integration with other chemistry software and applications.x
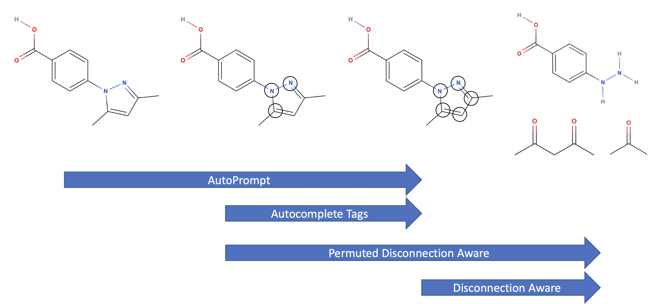
Figure 2: The disconnection-aware model is capable of four different prediction problems, depending on the granularity of control desired by the chemist.
Empirical results show that the use of disconnection prompts results in a 39% performance improvement over the baseline, as well as a 100% improvement in class diversity. This approach is effective in different chemistry domains, including traditional and enzymatic reactions. Overall, the use of disconnection prompts in retrosynthesis presents a promising approach to overcoming training data biases and improving the diversity of recommendations.
 >
> 

