Where AI May Be Used in Metabolomic Research: Data Processing
In the realm of metabolomics, where intricate networks of small molecules hold the keys to understanding biological systems, researchers face a constant battle against complexity. From optimizing analytical methods to tailoring sample preparation, the variables at play can overwhelm even the most seasoned scientists. But what if there was a way to harness the power of artificial intelligence (AI) to streamline experiment planning and unlock new frontiers of metabolomic discovery?
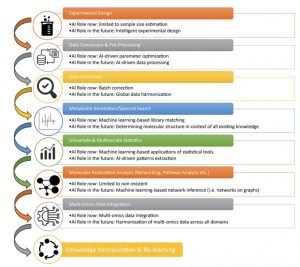
1. AI facilitates metabolite annotation by generating comprehensive in silico spectral libraries, improving spectral matching, and providing accurate molecular structure prediction.
The key challenge in metabolomics is translating spectral patterns to molecular identities. AI enhances the conventional approach of matching against reference libraries in multiple ways. It enables advanced similarity scoring accounting for noise, missing peaks, etc. by learning complex spectral relationships, instead of simplistic measures like dot products.
Additionally, AI drives breakthroughs in computational spectra generation to overcome limitations of currently small experimental libraries. Databases like METLIN and MassBank utilize AI to synthesize comprehensive libraries spanning vast chemical space. Finally, AI excels at the inverse problem of predicting molecular structures directly from spectra using self-supervised learning on large datasets, exemplified by tools like Sirius. When combined with a digital laboratory and life science analytics platform AI is a powerful research tool.
2. Cutting-edge AI allows integrating multi-omics data like metagenomics and metabolomics by modeling interconnected relationships across these disparate datasets.
While DNA provides a blueprint of microbial genes, metabolomics captures the functional molecular outputs synthesized by these microbes. However, combining these complementary omics data is challenging due to differences in size, structure, and underlying biology. Powerful AI methods like MMVec can infer conditional probabilities linking microbes to specific metabolites by identifying co-occurrence patterns using matrix factorization.
Such AI-driven multi-omics integration provides a systems view of microbial metabolism and overcomes limitations of simplistic correlation analysis which is confounded by data compositionality. Extending beyond the microbiome, AI strategies integrating transcriptomics, proteomics and metabolomics hold promise to map multi-scale mechanisms of metabolite biosynthesis across complex biological communities.
3. Network analysis approaches leverage AI to infer associations and patterns in mass spec data represented as networks.
The chemical similarity between mass spec features can be represented using network models, where nodes indicate analytes and edges connect those with related structures based on spectral patterns. AI techniques like graph neural networks, community detection, network propagation and embedding can then mine these data networks to uncover molecular associations and biochemical relationships.
This approach is founded on the premise that structural likeness reflected in the spectra implies functional relatedness. Extracting connectivity patterns in the networks using AI enables annotating molecular families and biochemical pathways directly from the spectral data in an unsupervised manner, with potential to delineate novel biochemistries from unknown molecules.
4. AI will transform the scientific process by dynamically incorporating new knowledge into learning models, enabling interactive AI assistants, and collaborative hypothesis generation powered by large biodata aggregators.
The AI revolution catalyzes a paradigm shift in knowledge creation and dissemination. Instead of the centuries-old model of publishing findings as static interpretations in papers, AI will facilitate an iterative, self-updating process. New evidence will be rapidly assimilated into knowledge bases which recursively retrain AI models, perpetually refining the understanding in a virtuous learning cycle.
Accompanying this transition, the format for accessing scientific knowledge may evolve from publications toward interactive AI assistants that contextualize insights based on accumulated data and postulations. Moreover, meta-aggregators of multimodal data sources will allow probing high-dimensional biological phenomena and machine-driven abductive reasoning, accelerating the generation of unifying hypotheses from big data. However, oversight on AI system inputs and thoroughness of model reasoning will be crucial to prevent propagating errors or harmful biases through the continuously expanding knowledgebase. While the full implementation of AI-driven experimental design in metabolomics is still an emerging field, the potential benefits are undeniable. By harnessing the power of AI, researchers could unlock new levels of precision, efficiency, and insight, accelerating the pace of metabolomic discoveries and paving the way for groundbreaking advancements in fields ranging from biomedicine to agriculture.
For biotech companies, the integration of AI into metabolomic research could offer a competitive edge, enabling more targeted and impactful research initiatives. However, it is crucial to approach this endeavor with a thoughtful and strategic mindset, ensuring that AI is implemented as a collaborative tool, which when combined with scientific data preservation tools complements and enhances human expertise, rather than replacing it. By striking the right balance between cutting-edge technology and scientific acumen, the metabolomic research community can unlock a future where the complexities of small molecule networks are unraveled with unprecedented clarity and precision.
Sources:
- Coler EA, Chen W, Melnik AV, Morton JT, Aksenov AA. Metabolomics in the era of artificial intelligence. Microbiota and Host. 2024;2(1).
 >
> 

